Modern Technology For Combating Bacterial Water Contamination
By J. Michael Trapp
Mobile-enabled geographic information systems (GIS) herald a new age for bacterial source tracking, allowing increased stakeholder involvement, more informed decision-making, and enhanced water quality.
Across the U.S., municipalities and industries are forced to deal with an increasing number of contaminated waterways. These impairments, including 303(d) listings and Total Maximum Daily Loads (TMDLs), stem from a variety of pollutants, including metals, sediments, organic chemicals, and trash. However, with each of these potential impairments presenting unique challenges, the most prevalent issue involves fecal indicator bacteria, which accounts for 13 percent of all impairments nationwide — or roughly 10,781 impairments. California is a prime example of the widespread nature of this issue. The state alone has 627 bacteria-impaired watersheds that comprise 26,000 square miles — or 16 percent of the state’s land area.
Bacterial contamination is not only most prevalent but also one of the most complex types of contamination for which to develop and implement plans to achieve permit compliance. This is a result of the diverse nature of bacterial sources and sinks and their ability to reproduce in varied environments. To further compound the issue, the bacteria used for regulatory compliance are not the organisms that directly pose the public health concerns that regulatory permits are designed to protect. Rather, they are indicators of pathogens that epidemiology studies have linked directly to human health concerns. As a result, there has been a growing focus on research and innovation to develop technologies and tools to help managers and planners identify bacteria pollution sources, assess their relative health risks, develop new advanced treatments, and deploy GIS tools to help effectively place treatment.
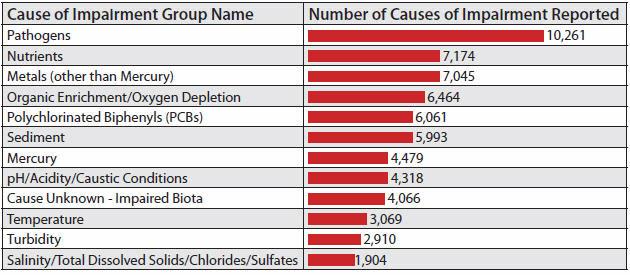
Causes of Impairment for 303(d) Listed Waters
Advances In Bacterial Source Tracking
Over the past decade, there have been significant advancements in the identification of sources of bacteria pollution in surface waters. Early efforts to reduce bacteria concentrations focused on wastewater discharges, as they frequently represent a major point source to a receiving waterbody that can be effectively managed. In many cases, the near-complete elimination of bacteria from these effluents was not sufficient to reach water quality goals, which left questions about the sources of the bacteria, particularly human sources, as they represent the greatest risk to public health.
To understand where the contamination is coming from, the field of microbial source tracking (MST) developed a toolbox of resources ranging from chemical tracers of human activities (such as optical brighteners or caffeine) to assays focused on the bacteria themselves. The earliest of these tools looked at ratios of fecal bacteria types and antibiotic resistance. More recent developments have provided genetic tools that are able to identify the specific host organisms of the bacteria and quantify an estimated contribution to the total load. The most common and widely used of the host-specific tests utilizes quantitative polymerase chain reaction (qPCR) technology to focus on the genetic code of the Bacteroides 16S rRNA gene. Small mutations in this gene have been found to be specific to populations of humans, birds (general avian, Canada geese, sea gulls, chickens), cows, pigs, horses, rodents, canines, and other common bacteria sources, which allow their discernment and quantification.
These tests have been successfully deployed in hundreds of source tracking programs around the country in either single-site temporal studies or across multiple watersheds to identify both host sources and geographical hot spots. This testing has been particularly successful in helping to identify failing septic tanks and leaking sewer infrastructure, as well as distinguishing other important sources to provide insight into their relative importance to the overall pollution load.
Quantitative Microbial Risk Assessment
Regulatory fecal indicator bacteria can come from many sources, representing a very different level of risk to public health. This is a result of the nature of the tests used, which focus on several species of indicator bacteria (e.g., E. coli, Enterococci, and fecal/total coliforms) as they are much easier to detect than the actual pathogens (bacterial and viral) that infect humans. While bacteria from human waste contains the highest concentration of human pathogens, and thus pose the largest threat for illness, other sources with high amounts of pathogens still pose varying risks to humans, including cow and canine waste and, to a lesser extent, bird waste.
Water quality standards were set based on the conservative principle that the indicator bacteria represent the worst-case scenario chance of infection. The protective manner in which these water quality standards were set means that in cases where impairment sources other than human are dominant, the same concentration of bacteria represents a different risk level. Recent research has also shown that many of these indicator bacteria can survive and reproduce in the environment and thus are not representative of new fecal pollution and pathogens inputs. Guidance from the EPA (2014) reflects this fact and allows a movement to bacteria standards that reflect rates of illness specific to a particular location.
Quantitative microbial risk assessment (QMRA) is the developing process that builds upon the understanding of site-specific source profiles determined in the MST process. QMRA can involve a number of steps, ranging from very precise sampling for determining pathogen concentrations to site-specific epidemiological studies. Data produced in this process are used to calculate a site-specific profile that determines an associated public health risk for different levels of bacterial concentrations. Based on this profile, a bacterial water quality standard can be set for that location that represents an acceptable illness rate and risk level.
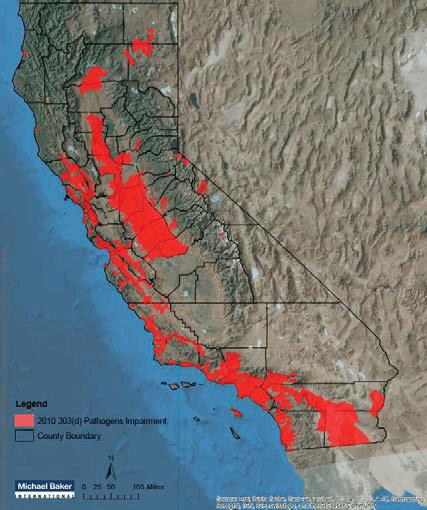
Map of 303(d) Pathogens Impairment in California (2010).
New Bacterial Treatment Options
While treatment of bacterial point sources is fairly well understood, nonpoint source bacteria present a more complicated problem. Source controls should always be the first and most effective choice, but they are not always sufficient to meet targets. Therefore, additional measures are often required.
Low-impact development (LID) and green infrastructure that emphasizes infiltration are preferred best management practices (BMP). These LID BMPs focus on the removal of flows carrying bacterial pollution and thus represent a powerful tool for remediation of bacterial pollution and offer the added benefits of hydromodification and groundwater replenishment. However, infiltration is not always feasible due to underling soils or volume requirement. As such, other types of structural treatments have been developed and utilized with varying levels of success. Early treatment attempted to mitigate in-stream bacteria by adding ozone and chlorine directly to impaired waters to kill the bacteria; however, these methods were often ineffective and often introduced additional problems to the watershed. More recently there have been a number of commercially available products that claim to treat bacteria, particularly in stormwater during the collection phase. Many of these interface directly with the catch basin structure. Examples include fabrics treated with antimicrobial elements that attach to floatable excluders and bioinfiltration media designed to kill bacteria by physically breaking down cell membranes.
GIS Innovation And Mobile Accessibility
The advancements in understanding the host sources, relative risks, and geographical hot spots have produced a wealth of knowledge for use in the decision-making process for dealing with bacterial pollution. That said, this data can be overwhelming if not organized in an easily digestible and relatable manner. Thankfully, the water and planning industries are in the midst of an information technology and mobile revolution. Today we are blessed with a number of desktop and mobile tools that focus on improving water quality and allow decision makers, policymakers, engineers, planners, and the general public to collaborate in an effort to achieve the shared goal of improving water quality in an impaired local watershed.
Central to this technological revolution are geospatial planning tools that build on GIS. Many different data layers are able to be observed simultaneously in the office and in the field on mobile devices to understand the interplay of sources with their surroundings. These new mobile-enabled GIS systems are designed to capture, store, manipulate, analyze, manage, and present a host of spatial and geographical data in real time. The available tools extend beyond accessibility to the technical elements, offering a look at the feasibility and costs of a watershed retrofit to help make informed decisions, apply for grants, and jumpstart long-term ideas based on current and future watershed concerns. iWATR (www.iwatr.com), for example, allows users to deeply understand their surrounding waterways by identifying potential sites for BMP, suggest BMP options for local impairment, estimate treatment volumes, and ultimately project costs for construction and lifetime maintenance and usage.
Summary
While bacterial contamination presents an important challenge to protecting the beneficial usages of our nation’s waters, there is an ever-growing toolkit of cutting-edge applications to meet them. If the past decade’s trend is any indication, technologies on the horizon will continue to revolutionize how we look at this problem. As technology advances and our reliance on mobile tools continues to increase, the GIS data available to all stakeholders will keep pace to help incite a more-educated population that understands the issues and works together to ensure we live in a world with clean and safe waterways.
About The Author
 Dr. J. Michael Trapp is a Project Manager for Michael Baker International in Southern California, focusing on water and sediment quality. He earned his Ph.D. from the University of Miami in marine and atmospheric chemistry and has experience conducting research on the fate and transport of aerosols, nutrients, metals, and bacterial contaminants.
Dr. J. Michael Trapp is a Project Manager for Michael Baker International in Southern California, focusing on water and sediment quality. He earned his Ph.D. from the University of Miami in marine and atmospheric chemistry and has experience conducting research on the fate and transport of aerosols, nutrients, metals, and bacterial contaminants.
